

The utility of a sequenced genome depends partially on the rate at which it has evolved. Many recent publications on Nematostella have been facilitated by this relational database. It also houses a primer database, a literature database and a database of living genetic stocks and DNA samples that is searchable by geographic locale or by population-specific genetic markers. The database is built on a relational structure in MySQL with a front-end HTML interface on an Apache server. StellaBase is a genomic database for Nematostella that allows users to search the genome sequence, predicted genes, predicted proteins cross-referenced with PFAM motifs ( 1) and expressed sequence tags (ESTs) ( 2).
/https://tf-cmsv2-photocontest-smithsonianmag-prod-approved.s3.amazonaws.com/4f91c7cbe78b2a8377e12e88cb4a03fd126efcb1.jpg)
The increasing utility of this species is evident from the recent surge in research papers-a PubMed search using the query ‘nematostella’ returns 35 journal articles published in 2006–07.
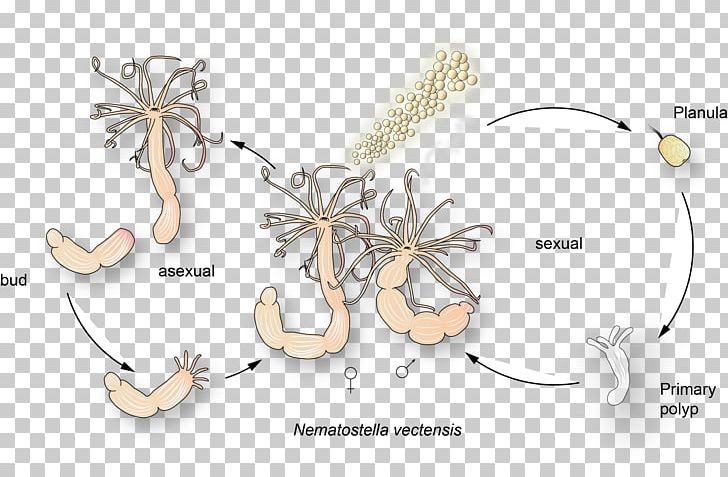
This species is emerging as an important model system in evolutionary genomics, developmental biology and estuarine ecology due to (i) the availability of a complete genome sequence, (ii) the generally conservative evolution of its genome, (iii) its ease of culture, (iv) the experimental tractability of its entire life history and (v) the ease of collecting the animal from the field. The starlet sea anemone, Nematostella vectensis, is a member of the basal metazoan phylum Cnidaria. StellaBase SNP ( ) is a relational database that describes the location and underlying type of mutation for 20 063 single nucleotide polymorphisms. We have developed two new databases that will foster such studies: StellaBase Disease ( ) is a relational database that houses 155 904 invertebrate homologous isoforms of human disease genes from four leading genomic model systems (fly, worm, yeast and Nematostella), including 14 874 predicted genes from the sea anemone itself. Recently, it has become apparent that Nematostella may be a particularly useful system for studying (i) microevolutionary variation in natural populations, and (ii) the functional evolution of human disease genes. StellaBase, the Nematostella Genomics Database ( ), was developed in 2005 as a resource to support the Nematostella research community. The starlet sea anemone, Nematostella vectensis, is a basal metazoan organism that has recently emerged as an important model system in developmental biology and evolutionary genomics.


 0 kommentar(er)
0 kommentar(er)
